Import EDAX EBSD Data (.h5)¶
Group (Subgroup)¶
Import/Export (Import)
Description¶
This Filter will read a single .h5 file into a new Data Container with a corresponding Image Geometry, allowing the immediate use of Filters on the data instead of having to generate the intermediate .h5ebsd file. A Cell Attribute Matrix and Ensemble Attribute Matrix will also be created to hold the imported EBSD information. Currently, the user has no control over the names of the created Attribute Arrays.
| User interface before entering a proper "Z Spacing" value and selecting which scans to include. |
|---|
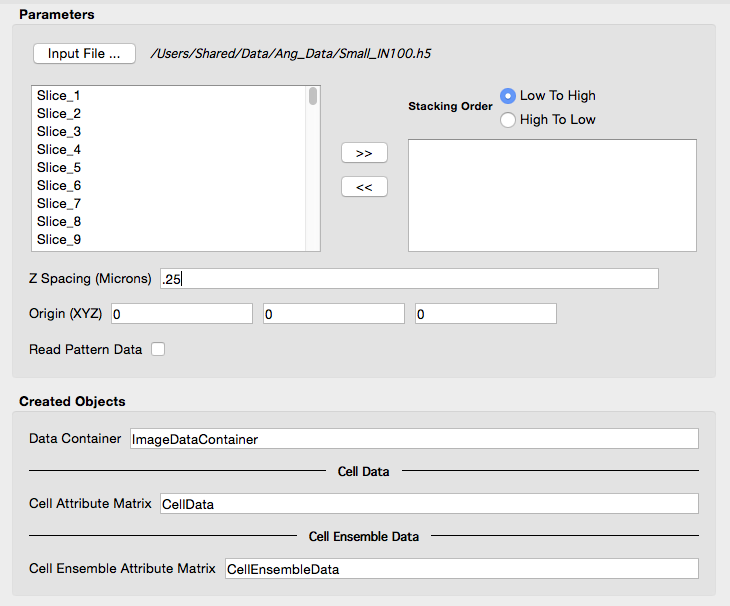 |
| User interface AFTER setting the "Z Spacing" and selecting files. |
|---|
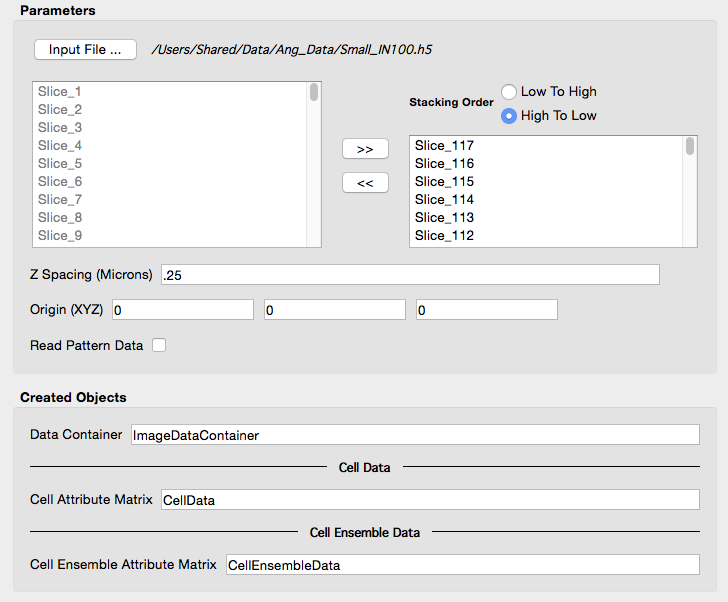 |
Notes About Reference Frames¶
The user should be aware that simply reading the file then performing operations that are dependent on the proper crystallographic and sample reference frame will be undefined or simply wrong. In order to bring the crystal reference frame and sample reference frame into coincidence, rotations will need to be applied to the data. The recommended filters are:
If the data has come from a TSL acquisition system and the settings of the acquisition software were in the default modes, the following reference frame transformations may need to be performed based on the version of the OIM Analysis software being used to collect the data:
- Sample Reference Frame: 180o about the <010> Axis
- Crystal Reference Frame: 90o about the <001> Axis
The user also may want to assign un-indexed pixels to be ignored by flagging them as "bad". The Threshold Objects Filter can be used to define this mask by thresholding on values such as Confidence Index > xx or Image Quality > desired quality.
Parameters¶
| Name | Type | Description |
|---|---|---|
| Input File | File Path | The input .h5 file path |
| Scan Name | Enumeration | The name of the scan in the .h5 file. EDAX can store multiple scans in a single file |
| Z Spacing | float | The spacing in microns between each layer. |
| Origin | float (3x1) | The origin of the volume |
| Read Pattern Data | bool | Default=OFF |
Required Geometry¶
Not Applicable
Required Objects¶
None
Created Objects¶
| Kind | Default Name | Type | Component Dimensions | Description |
|---|---|---|---|---|
| Data Container | ImageDataContainer | N/A | N/A | Created Data Container name with an Image Geometry |
| Attribute Matrix | CellData | Cell | N/A | Created Cell Attribute Matrix name |
| Attribute Matrix | CellEnsembleData | Cell Ensemble | N/A | Created Cell Ensemble Attribute Matrix name |
Created Cell Attribute Arrays¶
These arrays will most likely be created but is not guaranteed. Additional arrays (unknown at the time of writing) may also be created.
| Kind | Default Name | Type | Component Dimensions | Description |
|---|---|---|---|---|
| Cell Attribute Array | Confidence Index | float | (1) | Confidence of indexing |
| Cell Attribute Array | EulerAngles | float | (3) | Three angles defining the orientation of the Cell in Bunge convention (Z-X-Z) |
| Cell Attribute Array | Fit | float | (1) | Quality of fit for indexing |
| Cell Attribute Array | Image Quality | float | (1) | Quality of image |
| Cell Attribute Array | Phases | int32_t | (1) | Specifies to which phase each Cell belongs |
| Cell Attribute Array | SEM Signal | float | (1) | Value of SEM signal |
| Cell Attribute Array | X Position | float | (1) | X coordinate of Cell |
| Cell Attribute Array | Y Position | float | (1) | Y coordinate of Cell |
| Cell Attribute Array | Pattern | uint8_t | (NxM) | The pattern data may be very large. There is an option to NOT read it into DREAM.3D if it is not needed by the analysis. |
Created Ensemble Attribute Arrays¶
These arrays will most likely be created but is not guaranteed. Additional arrays (unknown at the time of writing) may also be created.
| Kind | Default Name | Type | Component Dimensions | Description |
|---|---|---|---|---|
| Ensemble Attribute Array | CrystalStructures | uint32_t | (1) | Enumeration representing the crystal structure for each Ensemble |
| Ensemble Attribute Array | LatticeConstants | float | (6) | The 6 values that define the lattice constants for each Ensemble |
| Ensemble Attribute Array | MaterialName | String | (1) | Name of each Ensemble |
Example Pipelines¶
License & Copyright¶
Please see the description file distributed with this Plugin
DREAM.3D Mailing Lists¶
If you need more help with a Filter, please consider asking your question on the DREAM.3D Users Google group!